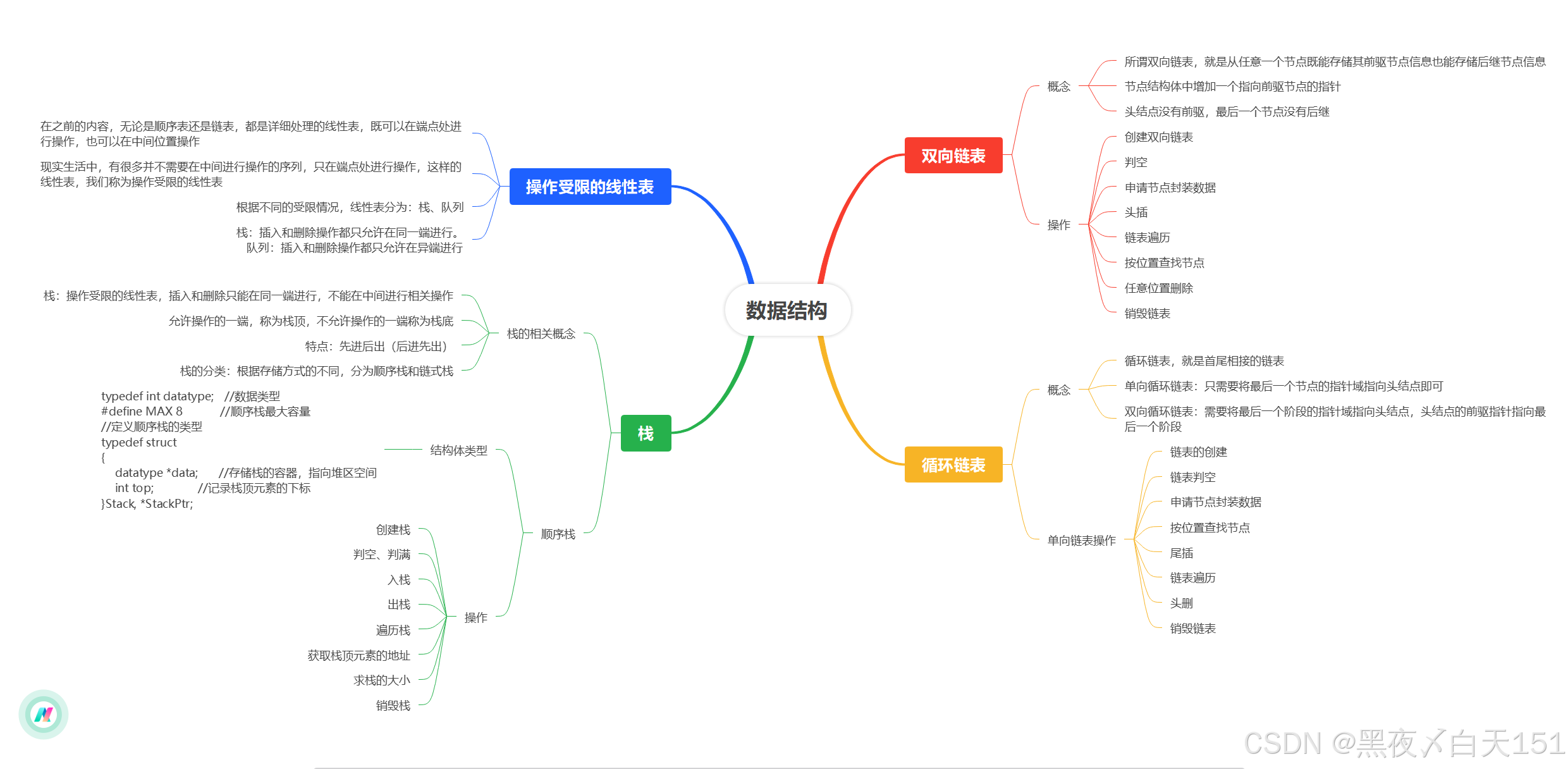
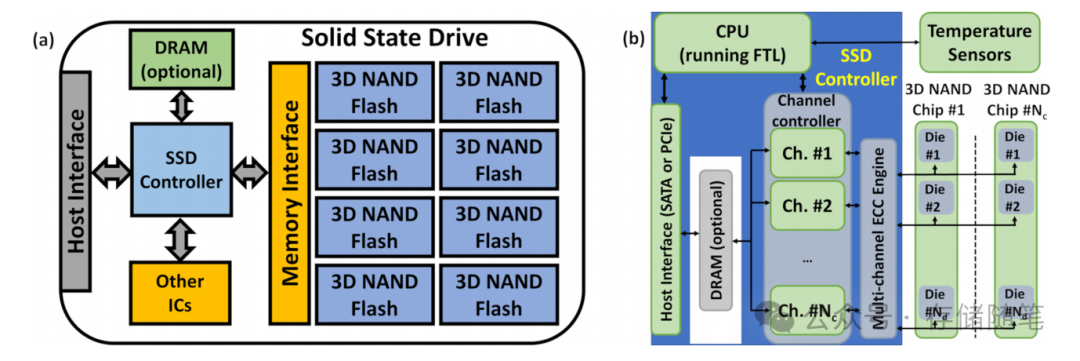
Visualizing the State-Space using the Continuous Wavelet Transform
import os
import pywt
#from wavelets.wave_python.waveletFunctions import *
import itertools
import numpy as np
import pandas as pd
from scipy.fftpack import fft
from collections import Counter
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from mpl_toolkits.axes_grid1 import make_axes_locatable
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib.patches as patchesLoad the el-Nino time-series dataset
dataset = "sst_nino3.dat.txt"
df_nino = pd.read_table(dataset)
N = df_nino.shape[0]
t0=1871
dt=0.25
time = np.arange(0, N) * dt + t0
signal = df_nino.values.squeeze()
print(time)
[1871. 1871.25 1871.5 1871.75 1872. 1872.25 1872.5 1872.75 1873.
1873.25 1873.5 1873.75 1874. 1874.25 1874.5 1874.75 1875. 1875.25
1875.5 1875.75 1876. 1876.25 1876.5 1876.75 1877. 1877.25 1877.5
1877.75 1878. 1878.25 1878.5 1878.75 1879. 1879.25 1879.5 1879.75
1880. 1880.25 1880.5 1880.75 1881. 1881.25 1881.5 1881.75 1882.
1882.25 1882.5 1882.75 1883. 1883.25 1883.5 1883.75 1884. 1884.25
1884.5 1884.75 1885. 1885.25 1885.5 1885.75 1886. 1886.25 1886.5
1886.75 1887. 1887.25 1887.5 1887.75 1888. 1888.25 1888.5 1888.75
1889. 1889.25 1889.5 1889.75 1890. 1890.25 1890.5 1890.75 1891.
1891.25 1891.5 1891.75 1892. 1892.25 1892.5 1892.75 1893. 1893.25
1893.5 1893.75 1894. 1894.25 1894.5 1894.75 1895. 1895.25 1895.5
1895.75 1896. 1896.25 1896.5 1896.75 1897. 1897.25 1897.5 1897.75
1898. 1898.25 1898.5 1898.75 1899. 1899.25 1899.5 1899.75 1900.
1900.25 1900.5 1900.75 1901. 1901.25 1901.5 1901.75 1902. 1902.25
1902.5 1902.75 1903. 1903.25 1903.5 1903.75 1904. 1904.25 1904.5
1904.75 1905. 1905.25 1905.5 1905.75 1906. 1906.25 1906.5 1906.75
1907. 1907.25 1907.5 1907.75 1908. 1908.25 1908.5 1908.75 1909.
1909.25 1909.5 1909.75 1910. 1910.25 1910.5 1910.75 1911. 1911.25
1911.5 1911.75 1912. 1912.25 1912.5 1912.75 1913. 1913.25 1913.5
1913.75 1914. 1914.25 1914.5 1914.75 1915. 1915.25 1915.5 1915.75
1916. 1916.25 1916.5 1916.75 1917. 1917.25 1917.5 1917.75 1918.
1918.25 1918.5 1918.75 1919. 1919.25 1919.5 1919.75 1920. 1920.25
1920.5 1920.75 1921. 1921.25 1921.5 1921.75 1922. 1922.25 1922.5
1922.75 1923. 1923.25 1923.5 1923.75 1924. 1924.25 1924.5 1924.75
1925. 1925.25 1925.5 1925.75 1926. 1926.25 1926.5 1926.75 1927.
1927.25 1927.5 1927.75 1928. 1928.25 1928.5 1928.75 1929. 1929.25
1929.5 1929.75 1930. 1930.25 1930.5 1930.75 1931. 1931.25 1931.5
1931.75 1932. 1932.25 1932.5 1932.75 1933. 1933.25 1933.5 1933.75
1934. 1934.25 1934.5 1934.75 1935. 1935.25 1935.5 1935.75 1936.
1936.25 1936.5 1936.75 1937. 1937.25 1937.5 1937.75 1938. 1938.25
1938.5 1938.75 1939. 1939.25 1939.5 1939.75 1940. 1940.25 1940.5
1940.75 1941. 1941.25 1941.5 1941.75 1942. 1942.25 1942.5 1942.75
1943. 1943.25 1943.5 1943.75 1944. 1944.25 1944.5 1944.75 1945.
1945.25 1945.5 1945.75 1946. 1946.25 1946.5 1946.75 1947. 1947.25
1947.5 1947.75 1948. 1948.25 1948.5 1948.75 1949. 1949.25 1949.5
1949.75 1950. 1950.25 1950.5 1950.75 1951. 1951.25 1951.5 1951.75
1952. 1952.25 1952.5 1952.75 1953. 1953.25 1953.5 1953.75 1954.
1954.25 1954.5 1954.75 1955. 1955.25 1955.5 1955.75 1956. 1956.25
1956.5 1956.75 1957. 1957.25 1957.5 1957.75 1958. 1958.25 1958.5
1958.75 1959. 1959.25 1959.5 1959.75 1960. 1960.25 1960.5 1960.75
1961. 1961.25 1961.5 1961.75 1962. 1962.25 1962.5 1962.75 1963.
1963.25 1963.5 1963.75 1964. 1964.25 1964.5 1964.75 1965. 1965.25
1965.5 1965.75 1966. 1966.25 1966.5 1966.75 1967. 1967.25 1967.5
1967.75 1968. 1968.25 1968.5 1968.75 1969. 1969.25 1969.5 1969.75
1970. 1970.25 1970.5 1970.75 1971. 1971.25 1971.5 1971.75 1972.
1972.25 1972.5 1972.75 1973. 1973.25 1973.5 1973.75 1974. 1974.25
1974.5 1974.75 1975. 1975.25 1975.5 1975.75 1976. 1976.25 1976.5
1976.75 1977. 1977.25 1977.5 1977.75 1978. 1978.25 1978.5 1978.75
1979. 1979.25 1979.5 1979.75 1980. 1980.25 1980.5 1980.75 1981.
1981.25 1981.5 1981.75 1982. 1982.25 1982.5 1982.75 1983. 1983.25
1983.5 1983.75 1984. 1984.25 1984.5 1984.75 1985. 1985.25 1985.5
1985.75 1986. 1986.25 1986.5 1986.75 1987. 1987.25 1987.5 1987.75
1988. 1988.25 1988.5 1988.75 1989. 1989.25 1989.5 1989.75 1990.
1990.25 1990.5 1990.75 1991. 1991.25 1991.5 1991.75 1992. 1992.25
1992.5 1992.75 1993. 1993.25 1993.5 1993.75 1994. 1994.25 1994.5
1994.75 1995. 1995.25 1995.5 1995.75 1996. 1996.25 1996.5 ]
# First lets load the el-Nino dataset, and plot it together with its time-average
def get_ave_values(xvalues, yvalues, n = 5):
signal_length = len(xvalues)
if signal_length % n == 0:
padding_length = 0
else:
padding_length = n - signal_length//n % n
xarr = np.array(xvalues)
yarr = np.array(yvalues)
xarr.resize(signal_length//n, n)
yarr.resize(signal_length//n, n)
xarr_reshaped = xarr.reshape((-1,n))
yarr_reshaped = yarr.reshape((-1,n))
x_ave = xarr_reshaped[:,0]
y_ave = np.nanmean(yarr_reshaped, axis=1)
return x_ave, y_ave
def plot_signal_plus_average(ax, time, signal, average_over = 5):
time_ave, signal_ave = get_ave_values(time, signal, average_over)
ax.plot(time, signal, label='signal')
ax.plot(time_ave, signal_ave, label = 'time average (n={})'.format(5))
ax.set_xlim([time[0], time[-1]])
ax.set_ylabel('Amplitude', fontsize=16)
ax.set_title('Signal + Time Average', fontsize=16)
ax.legend(loc='upper right')
fig, ax = plt.subplots(figsize=(12,3))
plot_signal_plus_average(ax, time, signal, average_over = 3)
plt.show()
Plot the Fourier Transform of the el-Nino dataset
def get_fft_values(y_values, T, N, f_s):
N2 = 2 ** (int(np.log2(N)) + 1) # round up to next highest power of 2
f_values = np.linspace(0.0, 1.0/(2.0*T), N2//2)
fft_values_ = fft(y_values)
fft_values = 2.0/N2 * np.abs(fft_values_[0:N2//2])
return f_values, fft_values
def plot_fft_plus_power(ax, time, signal, plot_direction='horizontal', yticks=None, ylim=None):
dt = time[1] - time[0]
N = len(signal)
fs = 1/dt
variance = np.std(signal)**2
f_values, fft_values = get_fft_values(signal, dt, N, fs)
fft_power = variance * abs(fft_values) ** 2
if plot_direction == 'horizontal':
ax.plot(f_values, fft_values, 'r-', label='Fourier Transform')
ax.plot(f_values, fft_power, 'k--', linewidth=1, label='FFT Power Spectrum')
elif plot_direction == 'vertical':
scales = 1./f_values
scales_log = np.log2(scales)
ax.plot(fft_values, scales_log, 'r-', label='Fourier Transform')
ax.plot(fft_power, scales_log, 'k--', linewidth=1, label='FFT Power Spectrum')
ax.set_yticks(np.log2(yticks))
ax.set_yticklabels(yticks)
ax.invert_yaxis()
ax.set_ylim(ylim[0], -1)
ax.legend()
fig, ax = plt.subplots(figsize=(12,3))
ax.set_xlabel('Frequency [Hz / year]', fontsize=18)
ax.set_ylabel('Amplitude', fontsize=18)
plot_fft_plus_power(ax, time, signal)
plt.show()
Plot the Scaleogram using the Continuous Wavelet Transform
def plot_wavelet(ax, time, signal, scales, waveletname = 'cmor',
cmap = plt.cm.seismic, title = '', ylabel = '', xlabel = ''):
dt = time[1] - time[0]
[coefficients, frequencies] = pywt.cwt(signal, scales, waveletname, dt)
power = (abs(coefficients)) ** 2
period = 1. / frequencies
levels = [0.0625, 0.125, 0.25, 0.5, 1, 2, 4, 8]
contourlevels = np.log2(levels)
im = ax.contourf(time, np.log2(period), np.log2(power), contourlevels, extend='both',cmap=cmap)
ax.set_title(title, fontsize=20)
ax.set_ylabel(ylabel, fontsize=18)
ax.set_xlabel(xlabel, fontsize=18)
yticks = 2**np.arange(np.ceil(np.log2(period.min())), np.ceil(np.log2(period.max())))
ax.set_yticks(np.log2(yticks))
ax.set_yticklabels(yticks)
ax.invert_yaxis()
ylim = ax.get_ylim()
ax.set_ylim(ylim[0], -1)
return yticks, ylim
scales = np.arange(1, 128)
title = 'Wavelet Transform (Power Spectrum) of signal'
ylabel = 'Period (years)'
xlabel = 'Time'
fig, ax = plt.subplots(figsize=(10, 10))
plot_wavelet(ax, time, signal, scales, xlabel=xlabel, ylabel=ylabel, title=title)
plt.show()
Plot all of the above in one figure
fig = plt.figure(figsize=(12,12))
spec = gridspec.GridSpec(ncols=6, nrows=6)
top_ax = fig.add_subplot(spec[0, 0:5])
bottom_left_ax = fig.add_subplot(spec[1:, 0:5])
bottom_right_ax = fig.add_subplot(spec[1:, 5])
plot_signal_plus_average(top_ax, time, signal, average_over = 3)
yticks, ylim = plot_wavelet(bottom_left_ax, time, signal, scales, xlabel=xlabel, ylabel=ylabel, title=title)
plot_fft_plus_power(bottom_right_ax, time, signal, plot_direction='vertical', yticks=yticks, ylim=ylim)
bottom_right_ax.set_ylabel('Period [years]', fontsize=14)
plt.tight_layout()
plt.show()
Empirical Wavelet Transform
#%% Example script
import numpy as np
import matplotlib.pyplot as plt
import ewtpy
T = 1000
t = np.arange(1,T+1)/T
noise = np.random.normal(0, 0.5, t.shape)
f = np.cos(2*np.pi*0.8*t) + 2*np.cos(2*np.pi*10*t)+0.8*np.cos(2*np.pi*100*t) + noise
ewt, mfb ,boundaries = ewtpy.EWT1D(f, N = 10)
plt.plot(f)
plt.plot(ewt)

Wavelet+LSTM

Using the Continuous Wavelet Transform and a Convolutional Neural Network for classification of signals

import pywt
import numpy as np
import matplotlib.pyplot as plt
from collections import defaultdict, Counter
import keras
from keras.layers import Dense, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras.models import Sequential
from keras.callbacks import History
history = History()
activities_description = {
1: 'walking',
2: 'walking upstairs',
3: 'walking downstairs',
4: 'sitting',
5: 'standing',
6: 'laying'
}
def read_signals(filename):
with open(filename, 'r') as fp:
data = fp.read().splitlines()
data = map(lambda x: x.rstrip().lstrip().split(), data)
data = [list(map(float, line)) for line in data]
return data
def read_labels(filename):
with open(filename, 'r') as fp:
activities = fp.read().splitlines()
activities = list(map(lambda x: int(x)-1, activities))
return activities
def randomize(dataset, labels):
permutation = np.random.permutation(labels.shape[0])
shuffled_dataset = dataset[permutation, :, :]
shuffled_labels = labels[permutation]
return shuffled_dataset, shuffled_labels
INPUT_FOLDER_TRAIN = 'data/train/InertialSignals/'
INPUT_FOLDER_TEST = 'data/test/InertialSignals/'
INPUT_FILES_TRAIN = ['body_acc_x_train.txt', 'body_acc_y_train.txt', 'body_acc_z_train.txt',
'body_gyro_x_train.txt', 'body_gyro_y_train.txt', 'body_gyro_z_train.txt',
'total_acc_x_train.txt', 'total_acc_y_train.txt', 'total_acc_z_train.txt']
INPUT_FILES_TEST = ['body_acc_x_test.txt', 'body_acc_y_test.txt', 'body_acc_z_test.txt',
'body_gyro_x_test.txt', 'body_gyro_y_test.txt', 'body_gyro_z_test.txt',
'total_acc_x_test.txt', 'total_acc_y_test.txt', 'total_acc_z_test.txt']
LABELFILE_TRAIN = 'data/train/y_train.txt'
LABELFILE_TEST = 'data/test/y_test.txt'
train_signals, test_signals = [], []
for input_file in INPUT_FILES_TRAIN:
signal = read_signals(INPUT_FOLDER_TRAIN + input_file)
train_signals.append(signal)
train_signals = np.transpose(np.array(train_signals), (1, 2, 0))
for input_file in INPUT_FILES_TEST:
signal = read_signals(INPUT_FOLDER_TEST + input_file)
test_signals.append(signal)
test_signals = np.transpose(np.array(test_signals), (1, 2, 0))
train_labels = read_labels(LABELFILE_TRAIN)
test_labels = read_labels(LABELFILE_TEST)
[no_signals_train, no_steps_train, no_components_train] = np.shape(train_signals)
[no_signals_test, no_steps_test, no_components_test] = np.shape(test_signals)
no_labels = len(np.unique(train_labels[:]))
print("The train dataset contains {} signals, each one of length {} and {} components ".format(no_signals_train, no_steps_train, no_components_train))
print("The test dataset contains {} signals, each one of length {} and {} components ".format(no_signals_test, no_steps_test, no_components_test))
print("The train dataset contains {} labels, with the following distribution:\n {}".format(np.shape(train_labels)[0], Counter(train_labels[:])))
print("The test dataset contains {} labels, with the following distribution:\n {}".format(np.shape(test_labels)[0], Counter(test_labels[:])))
uci_har_signals_train, uci_har_labels_train = randomize(train_signals, np.array(train_labels))
uci_har_signals_test, uci_har_labels_test = randomize(test_signals, np.array(test_labels))
The train dataset contains 7352 signals, each one of length 128 and 9 components
The test dataset contains 2947 signals, each one of length 128 and 9 components
The train dataset contains 7352 labels, with the following distribution:
Counter({5: 1407, 4: 1374, 3: 1286, 0: 1226, 1: 1073, 2: 986})
The test dataset contains 2947 labels, with the following distribution:
Counter({5: 537, 4: 532, 0: 496, 3: 491, 1: 471, 2: 420})Applying a CWT to UCI HAR signals and saving the resulting scaleogram into an numpy ndarray
scales = range(1,128)
waveletname = 'morl'
train_size = 5000
train_data_cwt = np.ndarray(shape=(train_size, 127, 127, 9))
for ii in range(0,train_size):
if ii % 1000 == 0:
print(ii)
for jj in range(0,9):
signal = uci_har_signals_train[ii, :, jj]
coeff, freq = pywt.cwt(signal, scales, waveletname, 1)
coeff_ = coeff[:,:127]
train_data_cwt[ii, :, :, jj] = coeff_
test_size = 500
test_data_cwt = np.ndarray(shape=(test_size, 127, 127, 9))
for ii in range(0,test_size):
if ii % 100 == 0:
print(ii)
for jj in range(0,9):
signal = uci_har_signals_test[ii, :, jj]
coeff, freq = pywt.cwt(signal, scales, waveletname, 1)
coeff_ = coeff[:,:127]
test_data_cwt[ii, :, :, jj] = coeff_Training a Convolutional Neural Network
x_train = train_data_cwt
y_train = list(uci_har_labels_train[:train_size])
x_test = test_data_cwt
y_test = list(uci_har_labels_test[:test_size])
img_x = 127
img_y = 127
img_z = 9
num_classes = 6
batch_size = 16
epochs = 10
# reshape the data into a 4D tensor - (sample_number, x_img_size, y_img_size, num_channels)
# because the MNIST is greyscale, we only have a single channel - RGB colour images would have 3
input_shape = (img_x, img_y, img_z)
# convert the data to the right type
x_train = x_train.reshape(x_train.shape[0], img_x, img_y, img_z)
x_test = x_test.reshape(x_test.shape[0], img_x, img_y, img_z)
#x_train = x_train.astype('float32')
#x_test = x_test.astype('float32')
print('x_train shape:', x_train.shape)
print(x_train.shape[0], 'train samples')
print(x_test.shape[0], 'test samples')
# convert class vectors to binary class matrices - this is for use in the
# categorical_crossentropy loss below
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)CNN
model = Sequential()
model.add(Conv2D(32, kernel_size=(5, 5), strides=(1, 1), activation='relu', input_shape=input_shape))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Conv2D(64, (5, 5), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten())
model.add(Dense(1000, activation='relu'))
model.add(Dense(num_classes, activation='softmax'))
model.compile(loss=keras.losses.categorical_crossentropy,
optimizer=keras.optimizers.Adam(),
metrics=['accuracy'])
model.fit(x_train, y_train, batch_size=batch_size,
epochs=epochs, verbose=1,
validation_data=(x_test, y_test),
callbacks=[history])
train_score = model.evaluate(x_train, y_train, verbose=0)
print('Train loss: {}, Train accuracy: {}'.format(train_score[0], train_score[1]))
test_score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss: {}, Test accuracy: {}'.format(test_score[0], test_score[1]))
fig, axarr = plt.subplots(figsize=(12,6), ncols=2)
axarr[0].plot(range(1, 11), history.history['acc'], label='train score')
axarr[0].plot(range(1, 11), history.history['val_acc'], label='test score')
axarr[0].set_xlabel('Number of Epochs', fontsize=18)
axarr[0].set_ylabel('Accuracy', fontsize=18)
axarr[0].set_ylim([0,1])
axarr[1].plot(range(1, 11), history.history['acc'], label='train score')
axarr[1].plot(range(1, 11), history.history['val_acc'], label='test score')
axarr[1].set_xlabel('Number of Epochs', fontsize=18)
axarr[1].set_ylabel('Accuracy', fontsize=18)
axarr[1].set_ylim([0.9,1])
plt.legend()
plt.show()
知乎学术咨询:https://www.zhihu.com/consult/people/792359672131756032?isMe=1工学博士,担任《Mechanical System and Signal Processing》审稿专家,担任《中国电机工程学报》优秀审稿专家,《控制与决策》,《系统工程与电子技术》,《电力系统保护与控制》,《宇航学报》等EI期刊审稿专家,擅长领域:现代信号处理,机器学习,深度学习,数字孪生,时间序列分析,设备缺陷检测、设备异常检测、设备智能故障诊断与健康管理PHM等。
知乎学术咨询:https://www.zhihu.com/consult/people/792359672131756032?isMe=1
工学博士,担任《Mechanical System and Signal Processing》审稿专家,担任《中国电机工程学报》优秀审稿专家,《控制与决策》,《系统工程与电子技术》,《电力系统保护与控制》,《宇航学报》等EI期刊审稿专家,擅长领域:现代信号处理,机器学习,深度学习,数字孪生,时间序列分析,设备缺陷检测、设备异常检测、设备智能故障诊断与健康管理PHM等。擅长领域:现代信号处理,机器学习,深度学习,数字孪生,时间序列分析,设备缺陷检测、设备异常检测、设备智能故障诊断与健康管理PHM等。