论文题目:ResViT: Residual vision transformers for multi-modal medical image synthesis
一种新的用于多模态医学图像合成的生成对抗方法

复现
1、从github下载源码
2、数据问题:
本次实验使用的数据不同于原项目,因此给出了新的dataset 方法和预处理方法
BraTS2020 Dataset (Training + Validation)
BraTS2020 Dataset (Training + Validation) (kaggle.com)
下载后解压
3、预处理,
预处理方法使用的MTNet的项目方法,代码讲解可以看我之前的博客
import numpy as np
from matplotlib import pylab as plt
import nibabel as nib
import random
import glob
import os
from PIL import Image
import imageio
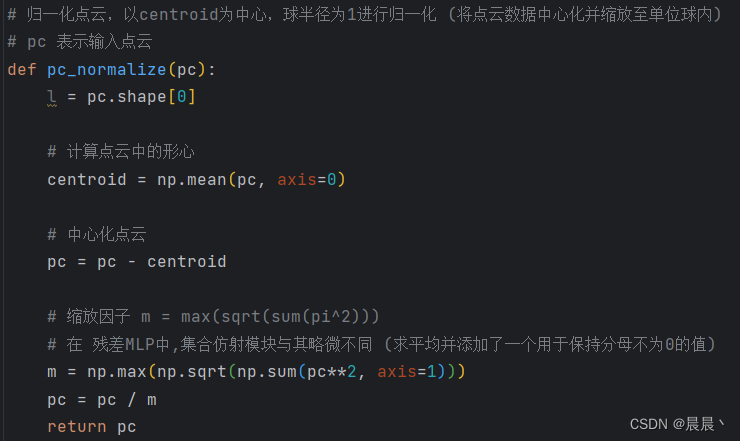
def normalize(image, mask=None, percentile_lower=0.2, percentile_upper=99.8):
if mask is None:
mask = image != image[0, 0, 0]
cut_off_lower = np.percentile(image[mask != 0].ravel(), percentile_lower)
cut_off_upper = np.percentile(image[mask != 0].ravel(), percentile_upper)
res = np.copy(image)
res[(res < cut_off_lower) & (mask != 0)] = cut_off_lower
res[(res > cut_off_upper) & (mask != 0)] = cut_off_upper
res = res / res.max() # 0-1
return res
def visualize(t1_data,t2_data,flair_data,t1ce_data,gt_data):
plt.figure(figsize=(8, 8))
plt.subplot(231)
plt.imshow(t1_data[:, :], cmap='gray')
plt.title('Image t1')
plt.subplot(232)
plt.imshow(t2_data[:, :], cmap='gray')
plt.title('Image t2')
plt.subplot(233)
plt.imshow(flair_data[:, :], cmap='gray')
plt.title('Image flair')
plt.subplot(234)
plt.imshow(t1ce_data[:, :], cmap='gray')
plt.title('Image t1ce')
plt.subplot(235)
plt.imshow(gt_data[:, :])
plt.title('GT')
plt.show()
def visualize_to_gif(t1_data, t2_data, t1ce_data, flair_data):
transversal = []
coronal = []
sagittal = []
slice_num = t1_data.shape[2]
for i in range(slice_num):
sagittal_plane = np.concatenate((t1_data[:, :, i], t2_data[:, :, i],
t1ce_data[:, :, i],flair_data[:, :, i]),axis=1)
coronal_plane = np.concatenate((t1_data[i, :, :], t2_data[i, :, :],
t1ce_data[i, :, :],flair_data[i, :, :]),axis=1)
transversal_plane = np.concatenate((t1_data[:, i, :], t2_data[:, i, :],
t1ce_data[:, i, :],flair_data[:, i, :]),axis=1)
transversal.append(transversal_plane)
coronal.append(coronal_plane)
sagittal.append(sagittal_plane)
imageio.mimsave("./transversal_plane.gif", transversal, duration=0.01)
imageio.mimsave("./coronal_plane.gif", coronal, duration=0.01)
imageio.mimsave("./sagittal_plane.gif", sagittal, duration=0.01)
return
if __name__ == '__main__':
t1_list = sorted(glob.glob(
'../data/BraTS2020_TrainingData/MICCAI_BraTS2020_TrainingData/*/*t1.*'))
t2_list = sorted(glob.glob(
'../data/BraTS2020_TrainingData/MICCAI_BraTS2020_TrainingData/*/*t2.*'))
t1ce_list = sorted(glob.glob(
'../data/BraTS2020_TrainingData/MICCAI_BraTS2020_TrainingData/*/*t1ce.*'))
flair_list = sorted(glob.glob(
'../data/BraTS2020_TrainingData/MICCAI_BraTS2020_TrainingData/*/*flair.*'))
gt_list = sorted(glob.glob(
'../data/BraTS2020_TrainingData/MICCAI_BraTS2020_TrainingData/*/*seg.*'))
data_len = len(gt_list)
train_len = int(data_len * 0.8)
test_len = data_len - train_len
train_path = '../data/train/'
test_path = '../data/test/'
os.makedirs(train_path,exist_ok=True)
os.makedirs(test_path,exist_ok=True)
for i,(t1_path, t2_path, t1ce_path, flair_path, gt_path) in enumerate(zip(t1_list,t2_list,t1ce_list,flair_list,gt_list)):
print('preprocessing the',i+1,'th subject')
t1_img = nib.load(t1_path) # (240,140,155)
t2_img = nib.load(t2_path)
flair_img = nib.load(flair_path)
t1ce_img = nib.load(t1ce_path)
gt_img = nib.load(gt_path)
#to numpy
t1_data = t1_img.get_fdata()
t2_data = t2_img.get_fdata()
flair_data = flair_img.get_fdata()
t1ce_data = t1ce_img.get_fdata()
gt_data = gt_img.get_fdata()
gt_data = gt_data.astype(np.uint8)
gt_data[gt_data == 4] = 3 #label 3 is missing in BraTS 2020
t1_data = normalize(t1_data) # normalize to [0,1]
t2_data = normalize(t2_data)
t1ce_data = normalize(t1ce_data)
flair_data = normalize(flair_data)
tensor = np.stack([t1_data, t2_data, t1ce_data, flair_data, gt_data]) # (4, 240, 240, 155)
if i < train_len:
for j in range(60):
Tensor = tensor[:, 10:210, 25:225, 50 + j]
np.save(train_path + str(60 * i + j + 1) + '.npy', Tensor)
else:
for j in range(60):
Tensor = tensor[:, 10:210, 25:225, 50 + j]
np.save(test_path + str(60 * (i - train_len) + j + 1) + '.npy', Tensor)
调整一下文件地址,处理好的如下

每一个npy都是一个 多模态切片 (5,200,200)
分别是t1_data, t2_data, t1ce_data, flair_data, gt_data
4、修改dataset
data/aligned_dataset.py
任务可以根据调整AB 修改
import os.path
import random
# import torchvision.transforms as transforms
import torch
from data.base_dataset import BaseDataset
# from data.image_folder import make_dataset
# from PIL import Image
import numpy as np
class AlignedDataset(BaseDataset):
def initialize(self, opt):
self.opt = opt
self.root = opt.dataroot
self.pathlist = os.listdir((self.root))
# self.dir_AB = os.path.join(opt.dataroot, opt.phase)
# self.AB_paths = sorted(make_dataset(self.dir_AB))
# assert(opt.resize_or_crop == 'resize_and_crop')
def __getitem__(self, index):
casepath = self.pathlist[index]
data = np.load(os.path.join(self.root,casepath))
# [t1_data, t2_data, t1ce_data, flair_data, gt_data]
A = data.take([0,1,3],0)
B = data[2,:,:]
B = np.expand_dims(B,0)
w_offset = random.randint(0, max(0, self.opt.loadSize - self.opt.fineSize - 1))
h_offset = random.randint(0, max(0, self.opt.loadSize - self.opt.fineSize - 1))
A = A[:, h_offset:h_offset + self.opt.fineSize, w_offset:w_offset + self.opt.fineSize]
B = B[:, h_offset:h_offset + self.opt.fineSize, w_offset:w_offset + self.opt.fineSize]
A = torch.tensor(A).float()
B = torch.tensor(B).float()
return {'A': A, 'B': B, 'A_paths': casepath, 'B_paths':casepath}
def __len__(self):
return len(self.pathlist)
def name(self):
return 'AlignedDataset'
5、运行!
nohup python -u train.py --dataroot yourdata --name tot1ce_pre_trained --gpu_ids 7 --batchSize 100 --model resvit_many --which_model_netG res_cnn --which_direction AtoB --lambda_A 100 --dataset_mode aligned --norm batch --pool_size 0 --output_nc 1 --input_nc 3 --loadSize 128 --fineSize 128 --niter 50 --niter_decay 50 --save_epoch_freq 5 --checkpoints_dir checkpoints/ --display_id 0 --lr 0.0002 >log_tot1ce_pre.log 2>&1&根据自己的实际情况调节dataroot、gpu、batchSize


![[<span style='color:red;'>代码</span><span style='color:red;'>复</span><span style='color:red;'>现</span>]Self-Attentive Sequential Recommendation](https://img-blog.csdnimg.cn/direct/ce09b3d9ac0f4d58a2d794a7e2fd1694.png)