介绍
生存分析的目的是分析某个时间点的“生存概率”是多少。基于这样的研究目的,需要提供生存数据,它是一种由不同的开始时间和结束时间组成的事件-时间的数据,比如在癌症研究领域,研究手术到死亡的过程、治疗到疾病进展等等。
在开展生存分析前,需要了解什么是删失(censored)。对于确定的事件,由于其他原因导致其出现无法记录、无法观察等等,这些都可以称为删失。“删失(censored)数据指在观察或试验中,由于人力或其他原因未能观察到所感兴趣的事件发生,因而得到的数据。”
knitr::include_graphics("./InputData/figures/Survival/survival_censor.png")
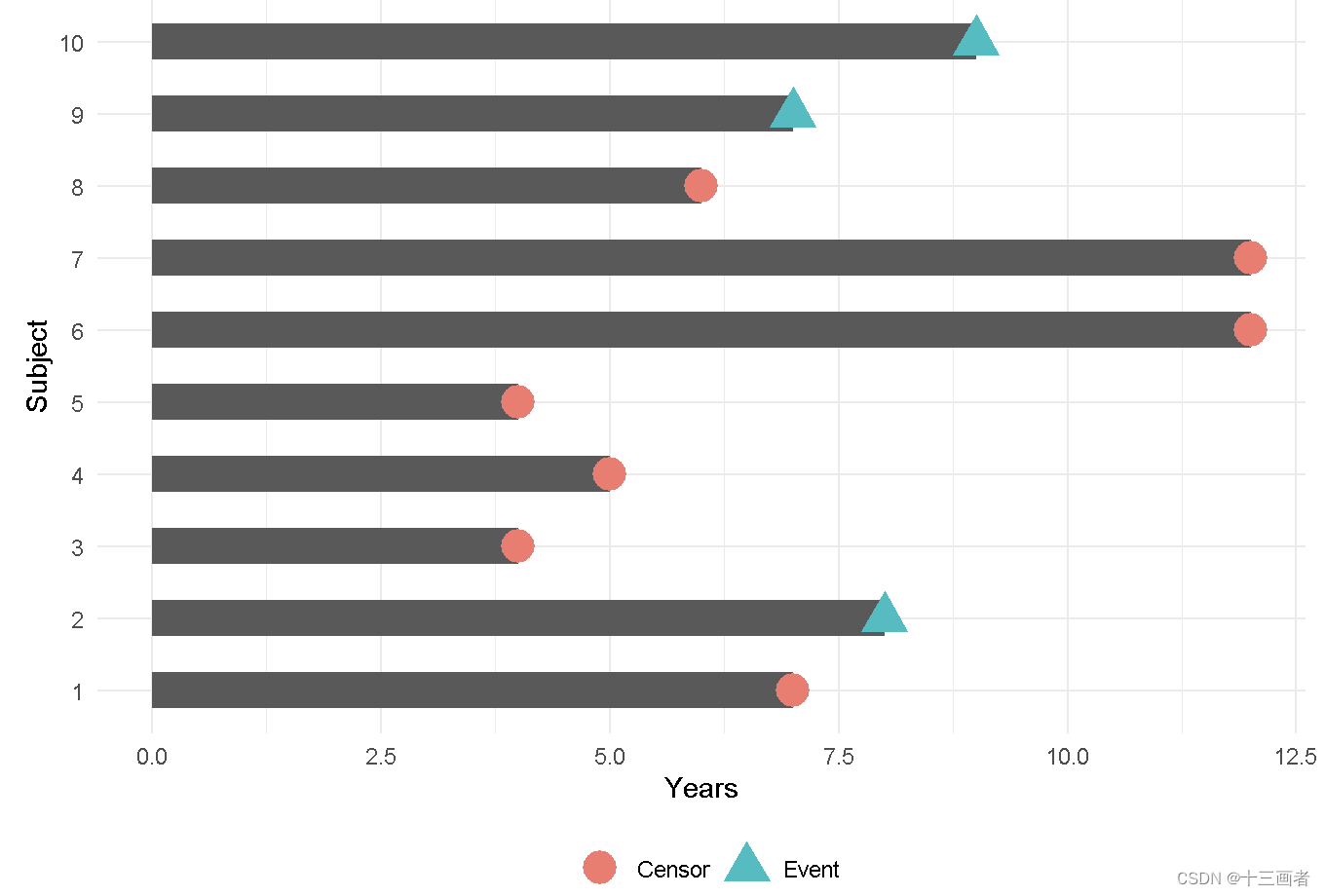
图表示10个参与者,在事件为发生前有7位患者出现了删失情况。忽略删失样本会导致生存概率结果出现偏差。生存分析是一种可以适当考虑被删失患者的方法。
组成
生存数据是有事件状态和对应时间组成,事件状态可以分成发生和删失。
事件时间: T i T_i Ti
删失时间: C i C_i Ci
事件状态:如果观察到事件则是1;否则是删失0。事件时间要小于删失时间。
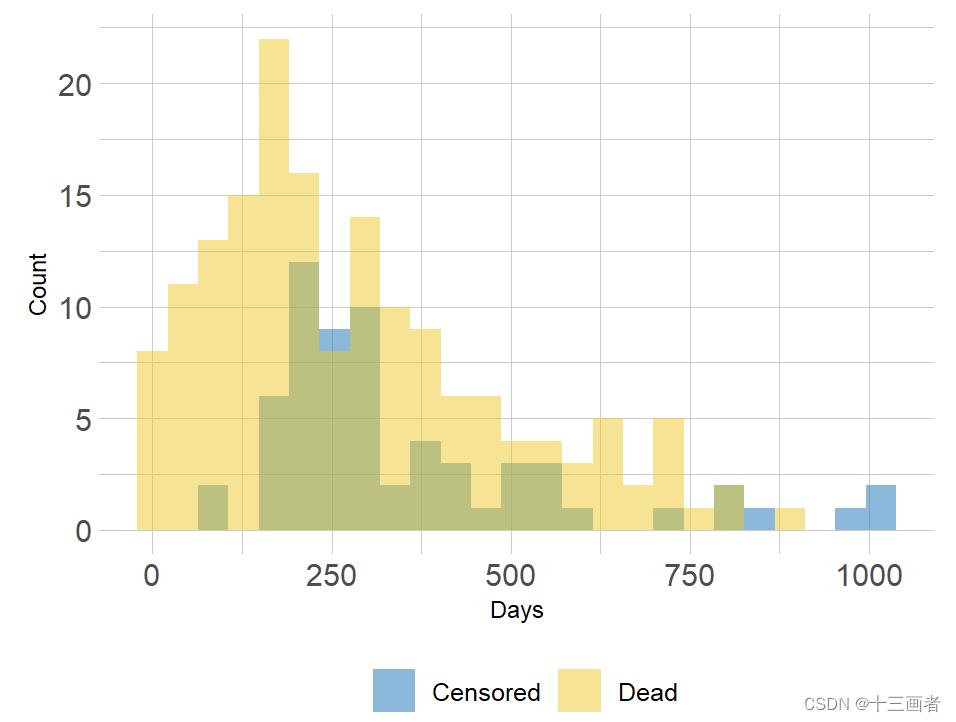
通过密度分布图可以观察到事件发生和删失状态在时间上的区别,如果不考虑删失则会导致评估结果偏高。
knitr::include_graphics("./InputData/figures/Survival/survival_event.png")
某个对象在某个时间点的生存概率公式为: S ( t ) = P r ( T > t ) = 1 − F ( t ) S(t) = Pr(T > t) = 1- F(t) S(t)=Pr(T>t)=1−F(t)
S ( t ) S(t) S(t)是生存函数
F ( t ) = 1 − P r ( T > t ) F(t) = 1- Pr(T > t) F(t)=1−Pr(T>t)是累积分布函数
案例
加载R包
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(tidyverse)
library(survival)
library(survminer)
library(gtsummary)
# rm(list = ls())
options(stringsAsFactors = F)
options(future.globals.maxSize = 1000 * 1024^2)
# group & color
sex_grp <- c("Male", "Female")
sex_col <- c("#F28880", "#60C4D3")
导入数据
肺癌数据: 本次研究目的----不同性别以及患者的生存状态是否存在差异
生存时间(time): 观察到的生存时间 (days)
生存状态(status): 死亡(2->1)和删失(1->0)
性别(sex): 1=Male, 2=Female
肿瘤活动状态(ph.ecog):0=ECOG0,1=ECOG1,2=ECOG2
dat <-
lung %>%
mutate(status = recode(status, `1` = 0, `2` = 1),
sex = recode(sex, `1` = "Male", `2` = "Female")) %>%
filter(!ph.ecog %in% c(NA, 3)) %>%
mutate(ph.ecog = recode(ph.ecog, `0` = "ECOG0", `1` = "ECOG1", `2` = "ECOG2"),
sex = factor(sex, levels = sex_grp))
head(dat[, c("time", "status", "sex", "ph.ecog")])

生存对象
通过Surv()生成生存对象,在使用survfit()生成生存曲线Kaplan-Meier plot的准备数据。
sur_fit <- survival::survfit(survival::Surv(time, status) ~ sex, data = dat)
sur_fit

组间生存时间差异比较
问题:在肺癌患者不同的性别分组中,生存概率是否存在差异性呢?
通常采用Log-rank检验推断两生存曲线整体间(整个观察期间)是否有差异,若生存曲线出现交叉情况,由于不满足成比例假设,则Log-rank检验不适用。
survminer::ggsurvplot(
fit = sur_fit,
pval = TRUE,
xlab = "Days",
ylab = "Overall survival probability",
legend.title = "Sex",
legend.labs = sex_grp,
palette = sex_col,
break.x.by = 100,
surv.median.line = "hv",
risk.table = TRUE,
risk.table.y.text = FALSE)
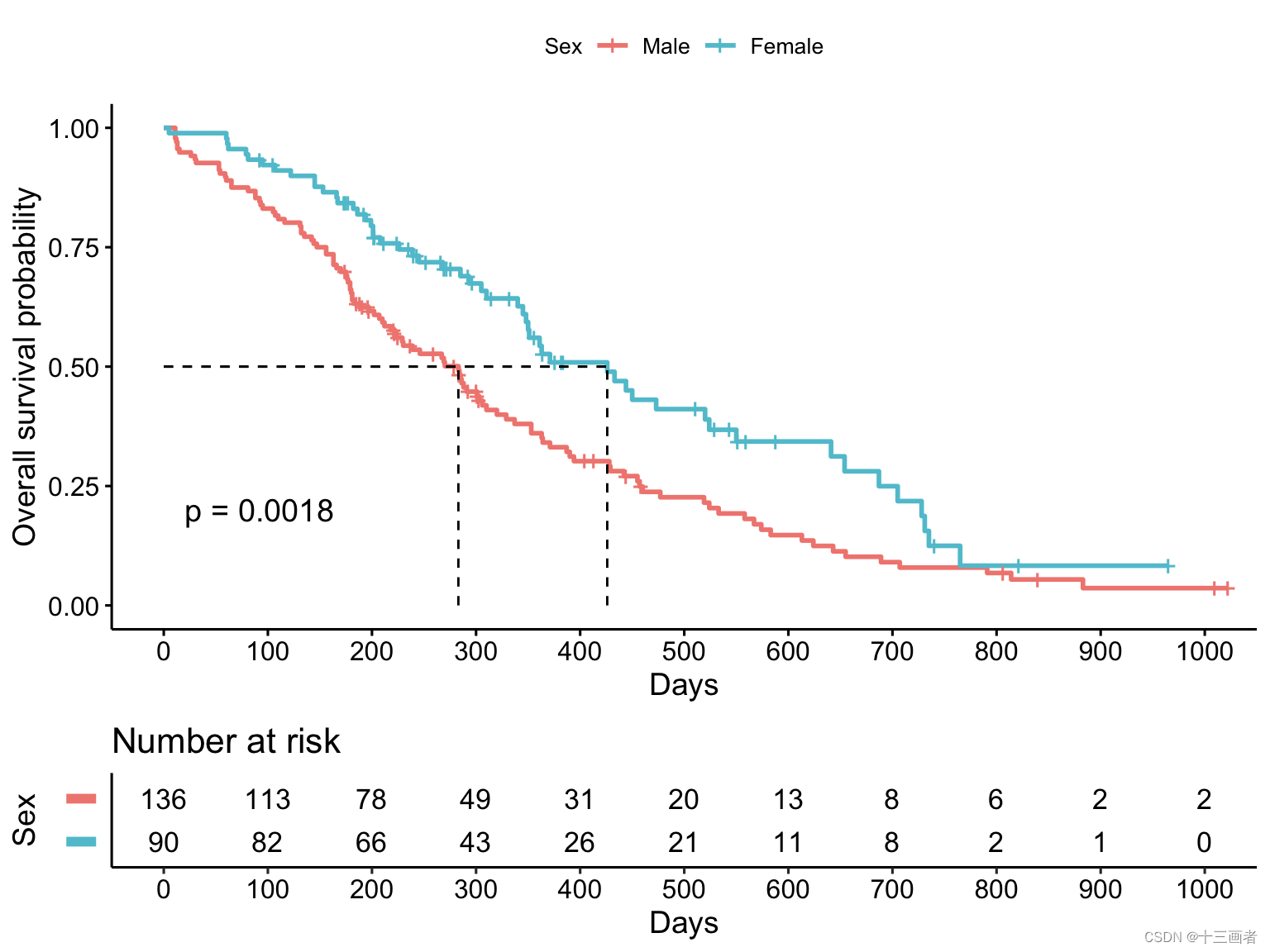
从图上的logRank p = 0.0018来看,女性在肺癌中生存周期更长。结合男性可能抽烟的额外因素考虑,该结果也符合预期。
中位生存期
中位生存期(Median Survival Time),又称为半数生存期, 即当累积生存率为0.5时所对应的生存时间,表示有且只有50%的个体可以活过这个时间。
sur_fit %>%
tbl_survfit(
probs = 0.5,
label_header = "**Median survival (95% CI)**"
)

女性的中位生存期(426天)要远远高于男性的(283天)
N年生存概率
选择1到3年生存概率,评估男女性间的差异
sur_fit %>%
tbl_survfit(
times = c(365, 730),
label_header = "**{time} Days survival (95% CI)**"
)
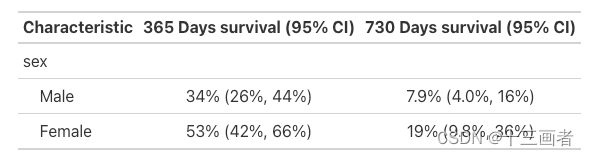
Cox回归模型
logRank test是定性变量是否和生存相关,定量变量之间的风险比例值需要用到Cox回归模型。单变量Cox回归仅评估该变量和生存的关系,多变量Cox回归可以校正其他因素影响后再评估单个变量的风险比例值。
broom::tidy(survival::coxph(Surv(time, status) ~ sex, data = dat)) %>% # conf.int = TRUE, exponentiate = TRUE
dplyr::select(-statistic) %>%
dplyr::mutate(`Hazard ratios` = exp(estimate),
HR_conf.low = exp(estimate) - 1.95*std.error,
HR_conf.high = exp(estimate) + 1.95*std.error,
HR = round(`Hazard ratios`, 2),
HR_conf.low = round(HR_conf.low, 2),
HR_conf.high = round(HR_conf.high, 2),
p.value = signif(p.value, 2)) %>%
dplyr::select(term, HR, HR_conf.low, HR_conf.high, p.value) %>%
rbind(as_tibble(data.frame(term = "RefMale",
HR = NA,
HR_conf.low = NA,
HR_conf.high = NA,
p.value = NA)))

以Male作为Reference,Female的风险比例值HR为0.6且对应的p<0.05,HR小于1,说明在肺癌患者中,Female的死亡风险要显著低于Male。
其他生存分析
探究不同性别分组的ECOG是否与生存状态相关
- 函数:1.获取生存分析结果;2.可视化生存曲线
get_cox_res <- function(
input, groups,
group_names, RefGroup) {
input_dat <- input %>%
dplyr::select(all_of(groups)) %>%
stats::setNames(c("TIME", "STATUS", "GROUP")) %>%
dplyr::filter(GROUP %in% group_names) %>%
dplyr::mutate(GROUP = factor(as.character(GROUP), levels = group_names))
## Group: log rank-test
fit <- survfit(Surv(TIME, STATUS) ~ GROUP, data = input_dat)
logrank <- surv_pvalue(fit = fit, data = input_dat)
## Group: Hazard ratios: Low as reference
HR <- broom::tidy(survival::coxph(survival::Surv(TIME, STATUS) ~ GROUP, data = input_dat)) %>%
dplyr::select(-statistic) %>%
dplyr::mutate(`Hazard ratios` = exp(estimate),
HR_conf.low = exp(estimate) - 1.95*std.error,
HR_conf.high = exp(estimate) + 1.95*std.error,
HR = signif(`Hazard ratios`, 2),
HR_conf.low = signif(HR_conf.low, 2),
HR_conf.high = signif(HR_conf.high, 2),
cox_pval = signif(p.value, 2),
HR_95 = paste0(HR, " (", HR_conf.low, " - ", HR_conf.high, ")")) %>%
dplyr::select(term, HR_95, cox_pval) %>%
rbind(data.frame(term = paste0("Ref", RefGroup),
HR_95 = NA,
cox_pval = NA))
## Group: Combination
res <- logrank %>%
dplyr::select(pval) %>%
dplyr::rename(logRank_pval = pval) %>%
rbind(data.frame(logRank_pval = rep(NA, (length(group_names) - 1) ))) %>%
cbind(HR) %>%
dplyr::select(term, logRank_pval, HR_95, cox_pval) %>%
dplyr::rename(logRank_pval = logRank_pval,
HR_95 = HR_95,
cox_pval = cox_pval)
return(res)
}
# survival plot
get_plot <- function(
input, groups, group_names,
feaID, TypeTime, NegGroup, PosGroup) {
input_dat <- input %>%
dplyr::select(all_of(groups)) %>%
stats::setNames(c("TIME", "STATUS", "GROUP")) %>%
dplyr::filter(GROUP %in% group_names) %>%
dplyr::mutate(GROUP = factor(as.character(GROUP), levels = group_names))
# survival model
fit <- survminer::surv_fit(Surv(TIME, STATUS) ~ GROUP, data = input_dat)
logrank <- surv_pvalue(fit = fit, data = input_dat)
pval_label <- paste(logrank$method, signif(logrank$pval, 3), sep = ": p=")
# median survival time
med_time <- fit %>%
gtsummary::tbl_survfit(
probs = 0.5,
label_header = "**Median survival (95% CI)**"
) %>%
as.tibble() %>%
dplyr::slice(-1) %>%
stats::setNames(c("Group", "Time")) %>%
dplyr::group_by(Group) %>%
dplyr::mutate(Median_temp = unlist(strsplit(Time, "\\s+\\("))[1]) %>%
dplyr::mutate(Median = ifelse(Median_temp == "-", "undef.", Median_temp),
Median_label = paste0(Group, " (", Median, " Days)")) %>%
dplyr::ungroup()
# cox model HR
cox_type <- rbind(NegGroup, PosGroup) %>%
dplyr::filter(!is.na(HR_95)) %>%
dplyr::mutate(term = gsub("GROUP", "", term)) %>%
dplyr::rename(Group = term) %>%
dplyr::mutate(HR_95 = gsub("\\s+-\\s+", ";", HR_95),
HR_95 = gsub("\\s+", "", HR_95),
cox_label = paste0("cox p=", cox_pval, ", HR=", HR_95),
logRank_pval = signif(logRank_pval, 3))
# merge labels
sur_labels <- med_time %>%
dplyr::left_join(cox_type %>%
dplyr::select(Group, cox_label),
by = "Group")
sur_labels$cox_label[is.na(sur_labels$cox_label)] <- "Reference"
sur_labels$final_label <- paste(sur_labels$Median_label, sur_labels$cox_label,
sep = " ")
# finale pvalue label
pval_label_final <- paste0(pval_label, "\n",
"(Male: p=", cox_type$logRank_pval[1],
"; ",
"Female: p=", cox_type$logRank_pval[2],
")")
ggsurv <- ggsurvplot(
fit,
pval = pval_label_final,
pval.size = 4,
pval.coord = c(0, 0.05),
xlab = "Overall Survival Time (Days)",
surv.scale = "percent",
break.time.by = 100,
risk.table.y.text.col = T,
risk.table = T,
risk.table.title = "",
risk.table.fontsize = 4,
risk.table.height = 0.3,
risk.table.y.text = FALSE,
ncensor.plot = FALSE,
surv.median.line = "hv",
palette = c("#803C08", "#F1A340",
"#2C0a4B", "#998EC3"),
legend = c(0.7, 0.89),
legend.labs = sur_labels$final_label,
legend.title = "",
font.legend = c(10, "plain"),
tables.theme = theme_cleantable(),
ggtheme = theme_classic() + theme(
axis.title = element_text(size = 10, face = "bold"),
axis.text = element_text(size = 9),
legend.background = element_rect(fill = "transparent", color = NA),
legend.box.background = element_rect(fill = "transparent", color = NA)
)
)
return(ggsurv)
}
- 准备数据和获取生存结果
dat_new <- dat %>%
dplyr::select(time, status, sex, ph.ecog) %>%
dplyr::mutate(MixedGroup = paste(sex, ph.ecog, sep = "_")) %>%
dplyr::mutate(MixedGroup = factor(MixedGroup,
levels = c("Male_ECOG0", "Male_ECOG1", "Male_ECOG2",
"Female_ECOG0", "Female_ECOG1", "Female_ECOG2")),
sex = factor(sex, levels = sex_grp))
MaleGroup <- get_cox_res(
input = dat_new,
groups = c("time", "status", "MixedGroup"),
group_names = c("Male_ECOG0", "Male_ECOG1"),
RefGroup = "Male_ECOG0")
FemaleGroup <- get_cox_res(
input = dat_new,
groups = c("time", "status", "MixedGroup"),
group_names = c("Female_ECOG0", "Female_ECOG1"),
RefGroup = "Female_ECOG0")
- 画图
ggsurpl <- get_plot(
input = dat_new,
groups = c("time", "status", "MixedGroup"),
group_names = c("Male_ECOG0", "Male_ECOG1",
"Female_ECOG0", "Female_ECOG1"),
feaID = "ECOG",
NegGroup = MaleGroup,
PosGroup = FemaleGroup)
ggsurpl
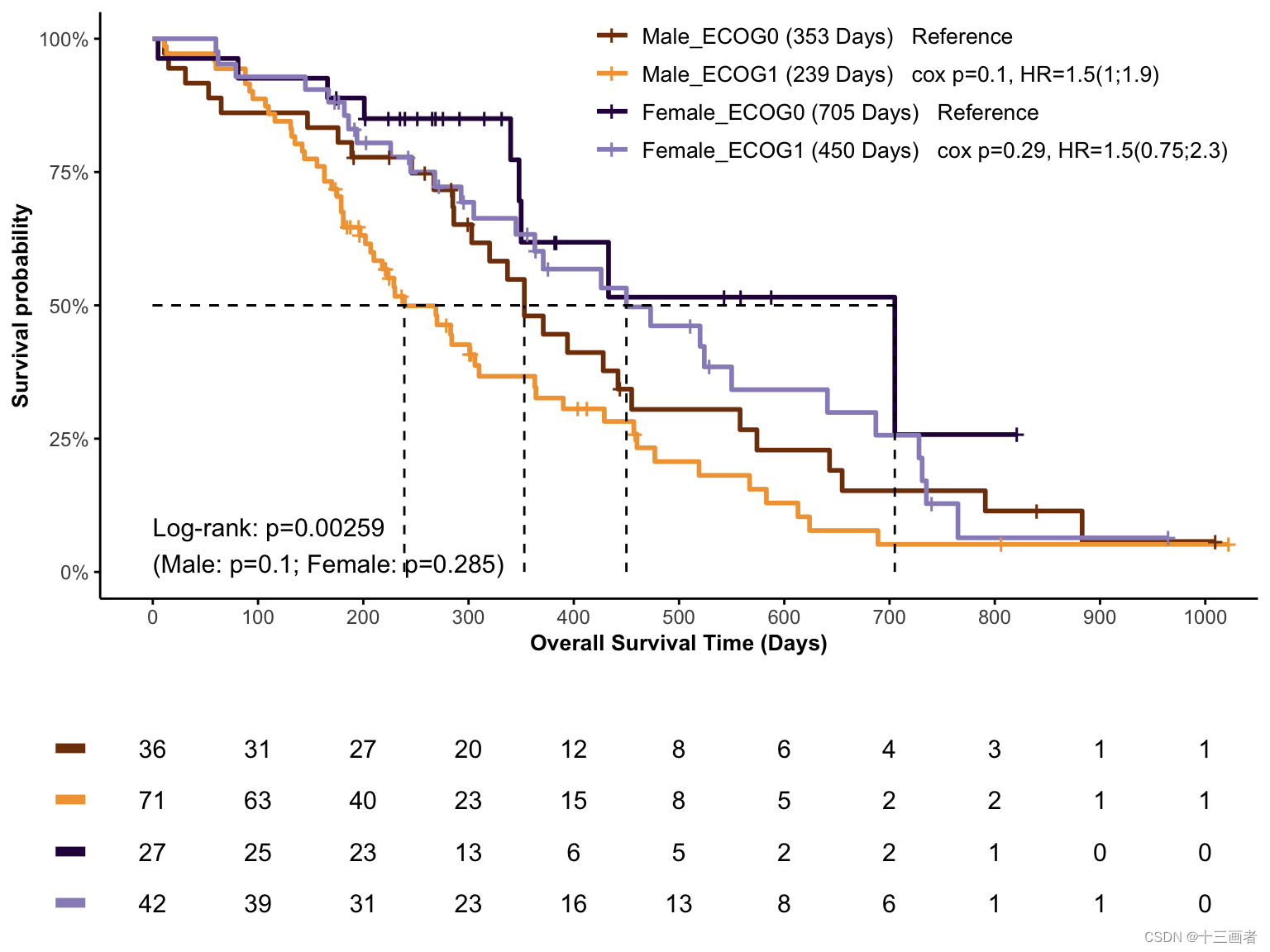
结果:不同性别分组内的ECOG组(ECOG0和ECOG1)与生存状态没有显著差异。
总结
生存数据很常见,是时间到事件的数据;
需要生存分析技术来解释删失的数据;
survival R包提供了生存分析工具,包括
Surv和surfit函数survminer R包提供了
ggsurvplot函数允许基于ggplot2定制Kaplan-Meier图组间比较可采用log-rank检验,采用
survival::survdiff单或多变量Cox回归分析可以使用
survival::Cox进行
Systemic information
devtools::session_info()
#> ─ Session info ───────────────────────────────────────────
#> setting value
#> version R version 4.3.1 (2023-06-16)
#> os macOS Monterey 12.2.1
#> system x86_64, darwin20
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Asia/Shanghai
#> date 2024-02-06
#> pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-5 2016-07-21 [1] CRAN (R 4.3.0)
#> backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
#> bookdown 0.37 2023-12-01 [1] CRAN (R 4.3.0)
#> broom 1.0.5 2023-06-09 [1] CRAN (R 4.3.0)
#> broom.helpers 1.14.0 2023-08-07 [1] CRAN (R 4.3.0)
#> bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.0)
#> cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
#> car 3.1-2 2023-03-30 [1] CRAN (R 4.3.0)
#> carData 3.0-5 2022-01-06 [1] CRAN (R 4.3.0)
#> cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.0)
#> colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
#> commonmark 1.9.1 2024-01-30 [1] CRAN (R 4.3.2)
#> data.table 1.15.0 2024-01-30 [1] CRAN (R 4.3.2)
#> devtools 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
#> digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.0)
#> downlit 0.4.3 2023-06-29 [1] CRAN (R 4.3.0)
#> dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.0)
#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
#> evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.0)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.0)
#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
#> fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
#> forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
#> fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
#> ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.3.0)
#> ggpubr * 0.6.0 2023-02-10 [1] CRAN (R 4.3.0)
#> ggsignif 0.6.4 2022-10-13 [1] CRAN (R 4.3.0)
#> ggtext 0.1.2 2022-09-16 [1] CRAN (R 4.3.0)
#> glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.0)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
#> gridtext 0.1.5 2022-09-16 [1] CRAN (R 4.3.0)
#> gt 0.10.1 2024-01-17 [1] CRAN (R 4.3.0)
#> gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
#> gtsummary * 1.7.2 2023-07-15 [1] CRAN (R 4.3.0)
#> highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
#> hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
#> htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.0)
#> htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.0)
#> httpuv 1.6.14 2024-01-26 [1] CRAN (R 4.3.2)
#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
#> jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.0)
#> km.ci 0.5-6 2022-04-06 [1] CRAN (R 4.3.0)
#> KMsurv 0.1-5 2012-12-03 [1] CRAN (R 4.3.0)
#> knitr 1.45 2023-10-30 [1] CRAN (R 4.3.0)
#> labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
#> later 1.3.2 2023-12-06 [1] CRAN (R 4.3.0)
#> lattice 0.21-8 2023-04-05 [1] CRAN (R 4.3.1)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.0)
#> lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.0)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
#> markdown 1.12 2023-12-06 [1] CRAN (R 4.3.0)
#> Matrix 1.6-5 2024-01-11 [1] CRAN (R 4.3.0)
#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
#> mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
#> miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
#> pkgbuild 1.4.3 2023-12-10 [1] CRAN (R 4.3.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
#> pkgload 1.3.4 2024-01-16 [1] CRAN (R 4.3.0)
#> profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.0)
#> promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.0)
#> purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
#> Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.0)
#> readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.0)
#> remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.3.0)
#> rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.0)
#> rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.0)
#> rstatix 0.7.2 2023-02-01 [1] CRAN (R 4.3.0)
#> rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
#> sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.0)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
#> shiny 1.8.0 2023-11-17 [1] CRAN (R 4.3.0)
#> stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.0)
#> stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.0)
#> survival * 3.5-5 2023-03-12 [1] CRAN (R 4.3.1)
#> survminer * 0.4.9 2021-03-09 [1] CRAN (R 4.3.0)
#> survMisc 0.5.6 2022-04-07 [1] CRAN (R 4.3.0)
#> tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
#> tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.2)
#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
#> tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
#> timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.0)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
#> urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
#> usethis 2.2.2 2023-07-06 [1] CRAN (R 4.3.0)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.0)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.0)
#> withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.0)
#> xfun 0.41 2023-11-01 [1] CRAN (R 4.3.0)
#> xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.0)
#> xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
#> yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.0)
#> zoo 1.8-12 2023-04-13 [1] CRAN (R 4.3.0)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/library
#>
#> ──────────────────────────────────────────────────────────